pytximport published
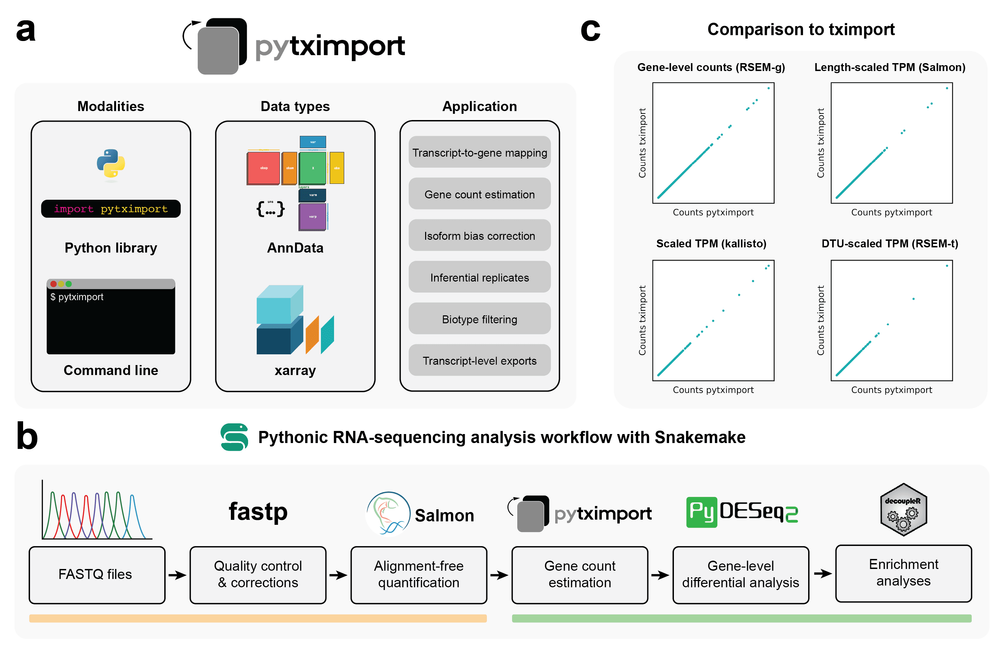
We are excited to announce the publication of our latest research paper in Bioinformatics, presenting pytximport, a Python implementation of the well-known tximport R package. This tool is designed to streamline transcript quantification in bulk RNA sequencing, addressing biases and enhancing compatibility with differential gene expression analysis tools in Python.
What is pytximport?
pytximport brings several powerful features to the table, including:
- Support for a variety of input formats.
- Multiple modes of bias correction.
- Handling inferential replicates.
- Gene-level summarization of transcript counts.
- Transcript-level exports.
- Generation of transcript-to-gene mappings.
- Optional filtering of transcripts by biotype.
Integration and Workflow
pytximport is part of the scverse ecosystem, a collection of open-source Python packages for omics analyses. It includes both a Python and command-line interface, making it versatile for different user preferences.
To demonstrate its utility, we have applied pytximport to a publicly available RNA-sequencing dataset, showcasing how it facilitates the creation of reproducible analysis workflows using Bioconda and scverse packages, all orchestrated through Snakemake rules.
Why pytximport?
By integrating pytximport into your analysis workflow, you can ensure more accurate and reliable results in RNA-seq data analysis, leveraging the robust tools provided by the scverse ecosystem.
We invite you to read our paper and explore pytximport to enhance your omics data analyses.